The thousands of diverse natural products made by Streptomyces species arise from simple organic building blocks, such as various amino acids, glucose-1-phosphate and malonyl-CoA. These primary metabolites are converted to complex natural products via multi-step biosynthetic pathways, which may include more than 30 individual steps. The first stage in a new project into the biosynthesis of a novel molecule is the identification of the genes responsible for its formation. This is facilitated by the fact that typically the genes associated for a given pathway are clustered together in the same locus. In order to find the biosynthetic genes, the genome of the producing Streptomyces species is first sequenced and putative gene clusters are identified by bioinformatics analysis. Next, experimental proof is required to demonstrate that the genes discovered are truly involved in the biosynthesis of the target metabolite. This can be done by gene inactivation experiments using CRISPR/Cas to generate a mutant Streptomyces strain where one of the genes in the gene cluster has been deleted. If the gene cluster is the correct one, then the strain should no longer be able to produce the target secondary metabolite and the biosynthesis comes to a halt at the stage where the gene product that was inactivated should function. In a complementation experiment, an intact copy of the same gene is re-introduced into the mutant strain, which should restore the ability of the strain to produce the natural product once again.
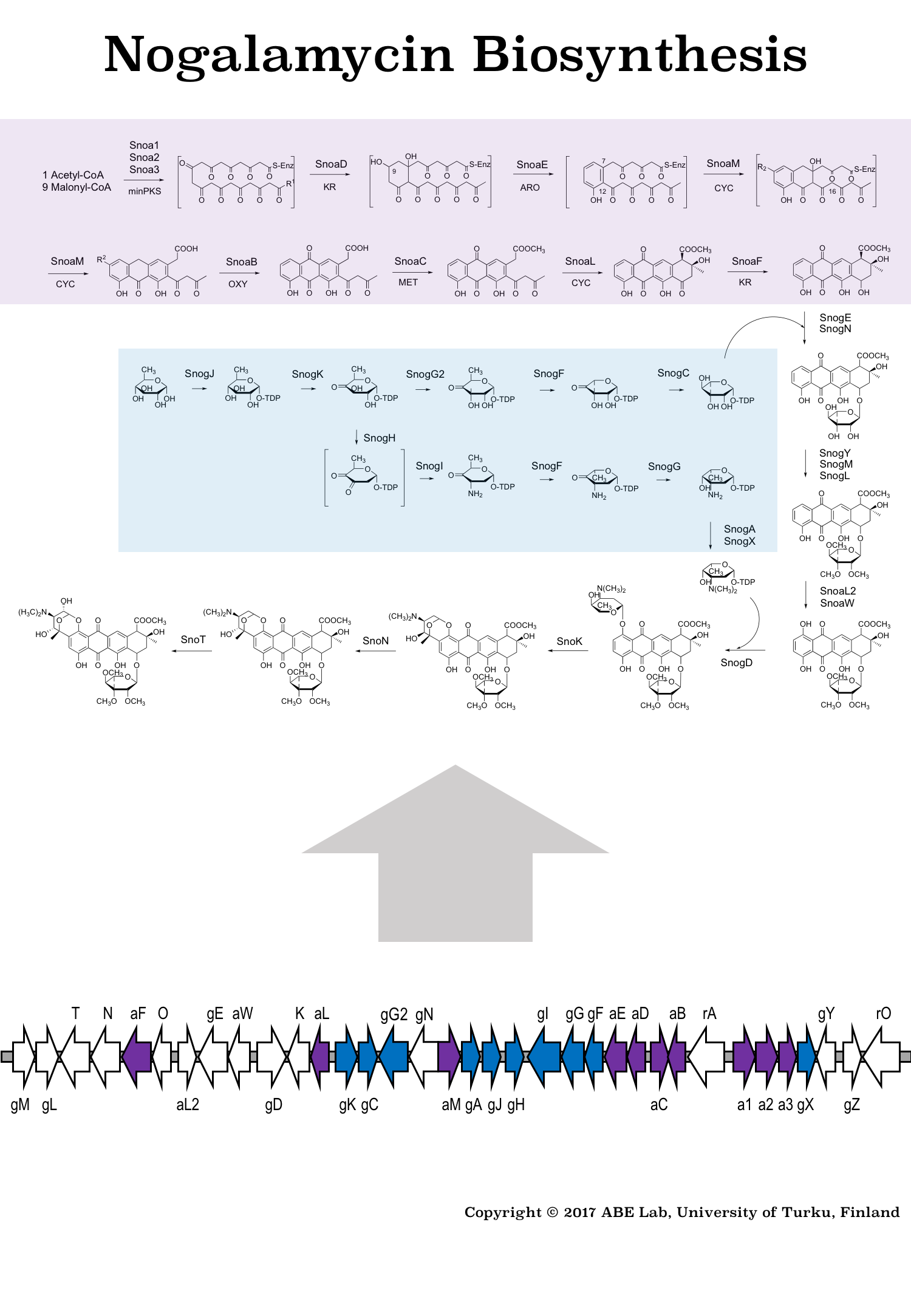
Figure 1: Biosynthetic pathway and gene cluster responsible for formation of the anti-cancer agent nogalamycin. The core structure of the natural product is made from acetyl- and malonyl-CoA building blocks and it is decorated with unusual carbohydrate units originating from glucose-1-phosphate.
Once the genes have been discovered, the biosynthesis can be studied in more detail by studying the proteins residing on the pathway. Key questions are the identification of the individual biosynthetic steps and determining the exact functions of the proteins on the pathway. The chemistry and structural diversity of many natural products is often unusual and unique, which is also reflected in the enzymes that are able to produce them. The study of these enzymes, the unusual catalysis they can perform, and the mechanisms by which catalysis is possible are often exciting and unexpected. Often, the first step in the identification of function involves sequence comparisons with proteins of known or putative function. From these initial steps, much information can be gained from extensive sequence data bases. This includes identifying motifs indicating the use of particular cofactors or identifying highly conserved amino acids which may reflect the important role they play within the structure or the catalytic machinery of the enzyme. Next enzymatic activity needs to be observed under laboratory conditions. This can be done by conducting enzymatic reactions and subsequent analysis of substrates and products by HPLC.
The detection of enzymatic activity is important, since this unequivocally confirms the function of the protein. In addition, this allows us to initiate even more detailed studies of the protein at the molecular level, which aim to understand the catalytic mechanism. How does the enzyme catalyze the reaction it does? The determination of the three dimensional structure of the protein by X-ray crystallography will provide a detailed picture of what the protein looks like and this vast information can be used to design functional studies. However, predicting the effects of mutations on catalysis can be challenging even when targeting a single amino acid in the active site and having the structure of the protein available. In the biosynthesis of secondary metabolites, we regularly encounter homologous enzymes with highly similar sequences and structures. Yet, despite their resemblances in both sequence and structure these enzymes can catalyze different reactions. The activities of the variants can then be measured to determine how the changes have influenced catalysis and used to decipher the different catalytic steps that occur during the reaction cycle. For these reasons, the assignment of function should not be based on sequence alignments or the structure alone, but should also require activity measurements.
Once all of this information has been gathered, we have acquired a very detailed picture of how the enzyme works. This allows us to ask one final question: can the enzyme be modified by protein engineering in such a way that it would catalyze completely novel chemistry? Incorporation of such proteins to pre-existing metabolic pathways could lead to the formation of novel antibiotics. We can use the atomic resolution structure of the protein and information from related enzymes to design changes in the active site to alter substrate binding or even catalysis. Alternatively, we can utilize directed evolution to generate a large library of mutants and high-throughput screening to find a variant that catalyzes the reaction we are interested in.
The ABELAB has expertise in all of these research areas. We are able to start working on previously unknown molecule, clone and manipulate the biosynthetic pathway and conduct investigation at the molecular level to understand the key steps in the biosynthesis. Student projects are typically available for all stages.
Updated: May 2018