How are Streptomyces bacteria able to produce tens of thousands of bioactive compounds? Many of these are chemically so complex that humans are not able to use organic chemistry to accomplish the same feat. The trick appears to lie in the rapid evolution of secondary metabolism, where even subtle changes in the biosynthetic enzymes may lead to formation of novel functions. Since the appearance of a novel bioactive compound may provide a significant competitive advantage to the producing organism, a strong evolutionary pressure exits that promotes formation of chemical diversity. Due to this phenomenon we have regularly encountered homologous enzymes with highly similar sequences and structures, which nonetheless catalyze different reactions.
As an example, in the biosynthesis of various anthracycline antibiotics we have discovered proteins that belong to the family of S-adenosyl-methionine (SAM) -dependent methyltransferases, but which catalyze either 4-O-methylation, 10-hydroxylation or 10-decarboxylation. In a typical evolutionary study, a phylogenetic tree is first constructed in order to obtain an overview of the evolutionary relationships of the proteins. This facilitates further analyses, since typically proteins are distributed into distinct clades based on their function, as was the case with the SAM-dependent anthracycline tailoring proteins.
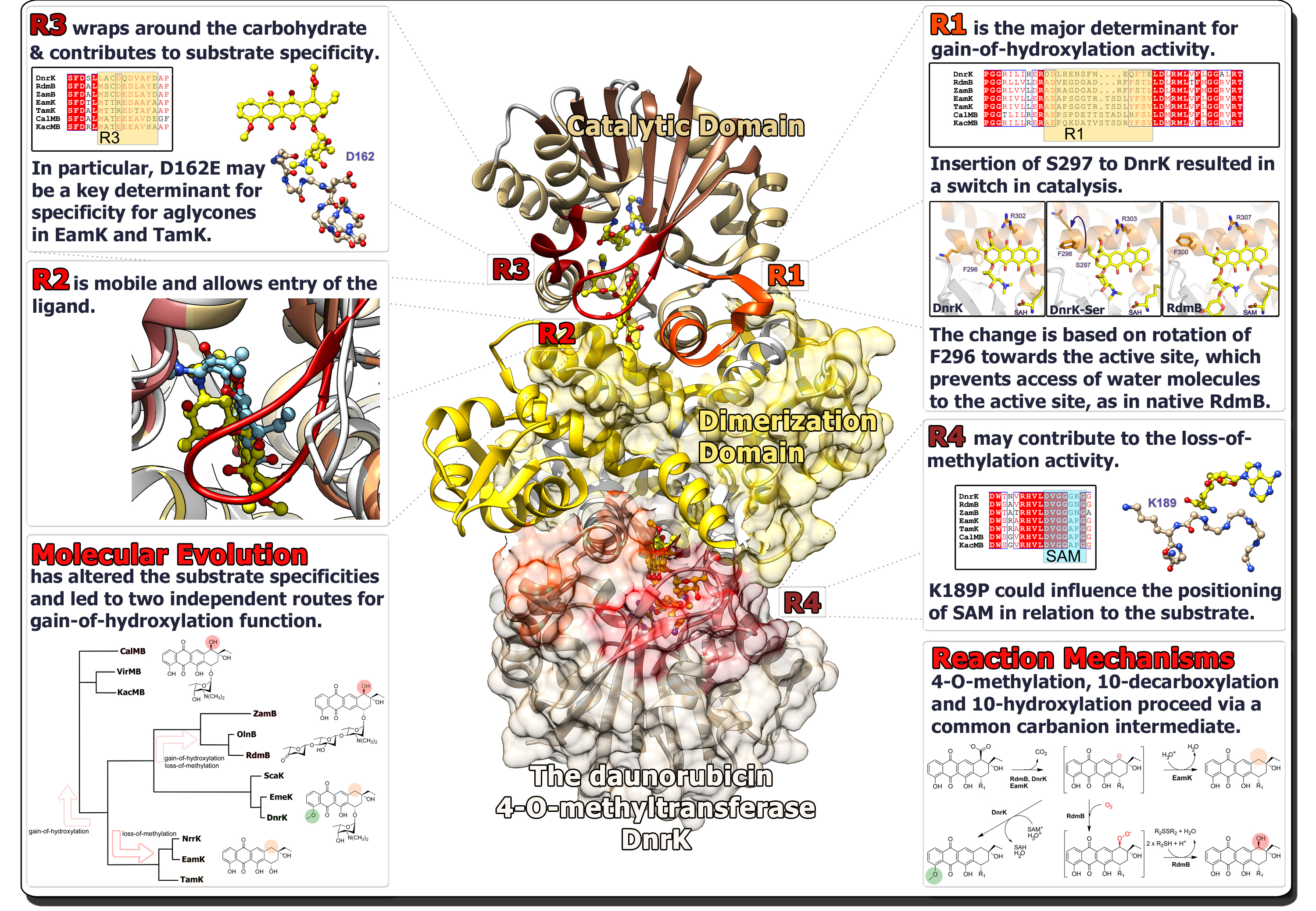
Since we often have crystal structures of the enzymes available, we are able to probe the molecular evolution of proteins in much more detail. Structural analysis may reveal evolutionary hotspots that are responsible for diversification of protein function. We have extensively utilized a protein engineering method called chimeragenesis, where several amino acids are exchanged between two homologous proteins, to modulate the activity of the enzymes. These regions may then be narrowed down to individual amino acids and structure determination of chimeric enzymes may then be used to probe the molecular basis for the switch in activity. Using such an approach, we were able to demonstrate that introduction of a single amino acid to the 4-O-methyltransferase DnrK resulted in the emergence of the 10-hydroxylation activity of RdmB. The amino acid insertion was located in an a16-helix adjacent to the active site, but in such a manner that the side chain was pointing away from the substrate. However, this led to rotation of a preceding phenylalanine towards the active site cavity, which enables the hydroxylation reaction to occur by blocking a water channel towards the surface of the enzyme.
These studies have relied on our incisive understanding of enzymes involved in the biosynthesis of secondary metabolites. We hope that when the individual constraints are known for each enzyme, we will be able to fully exploit protein engineering for designing potentially ground breaking drugs. The studies also provide fascinating insight into how enzymes are able to acquire novel functions during the course of evolution. The diversity of Streptomyces secondary metabolic pathways provide an exceptionally rich playground for such investigations.
Updated: November 2018