We sequence genomes and transcriptomes in various projects, e.g. secondary metabolite prediction, cluster activation and differential expression analysis, in order to understand the biosynthesis of natural products. Through next generation sequencing (NGS) technology, we are able to identify potential biosynthetic gene clusters (BGC), their regulatory elements and key enzymes to use as a basis for synthetic biology applications.
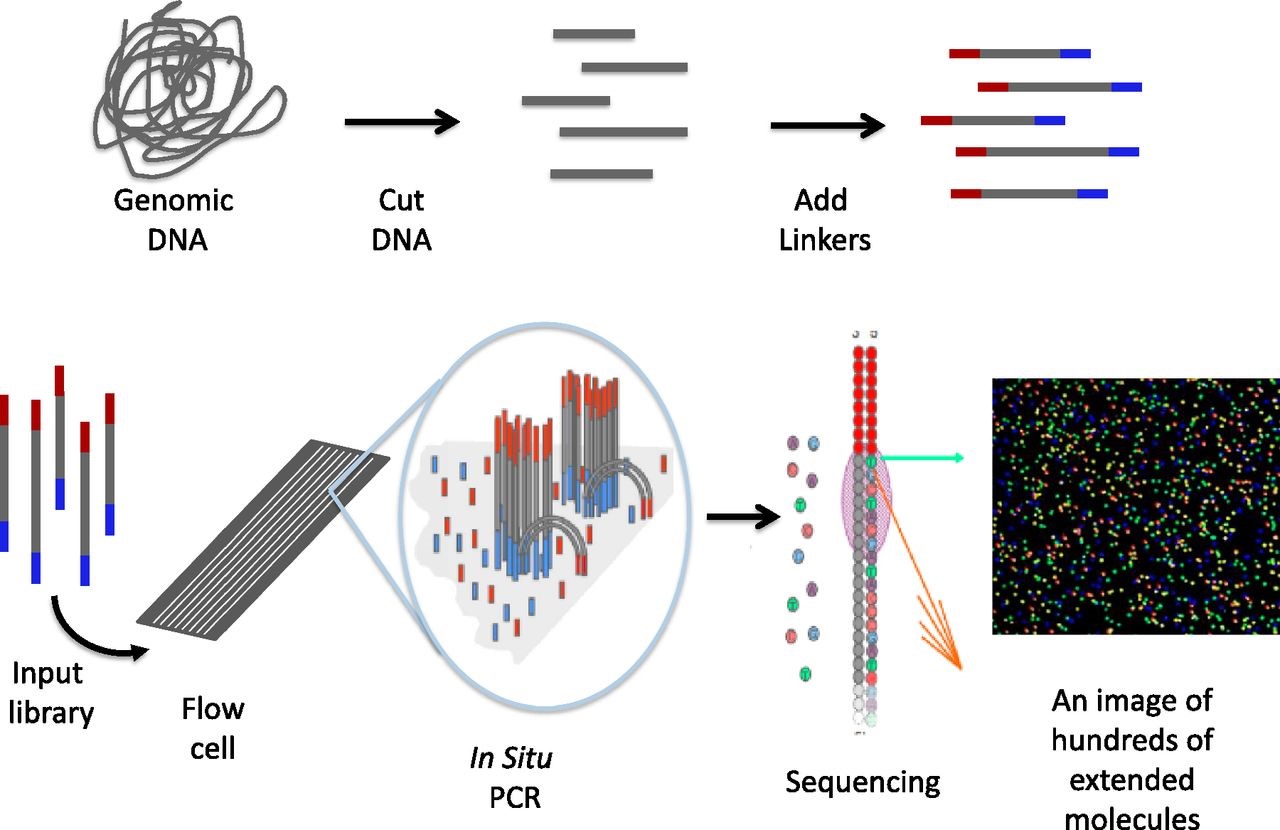
Figure 1. An overview of a general NGS workflow. (source)
We use the core facilities at Biocity to do our sequencing using the Illumina platform (HiSeq or MiSeq depending on our needs). The data analysis is done using the supercomputer provided by CSC.
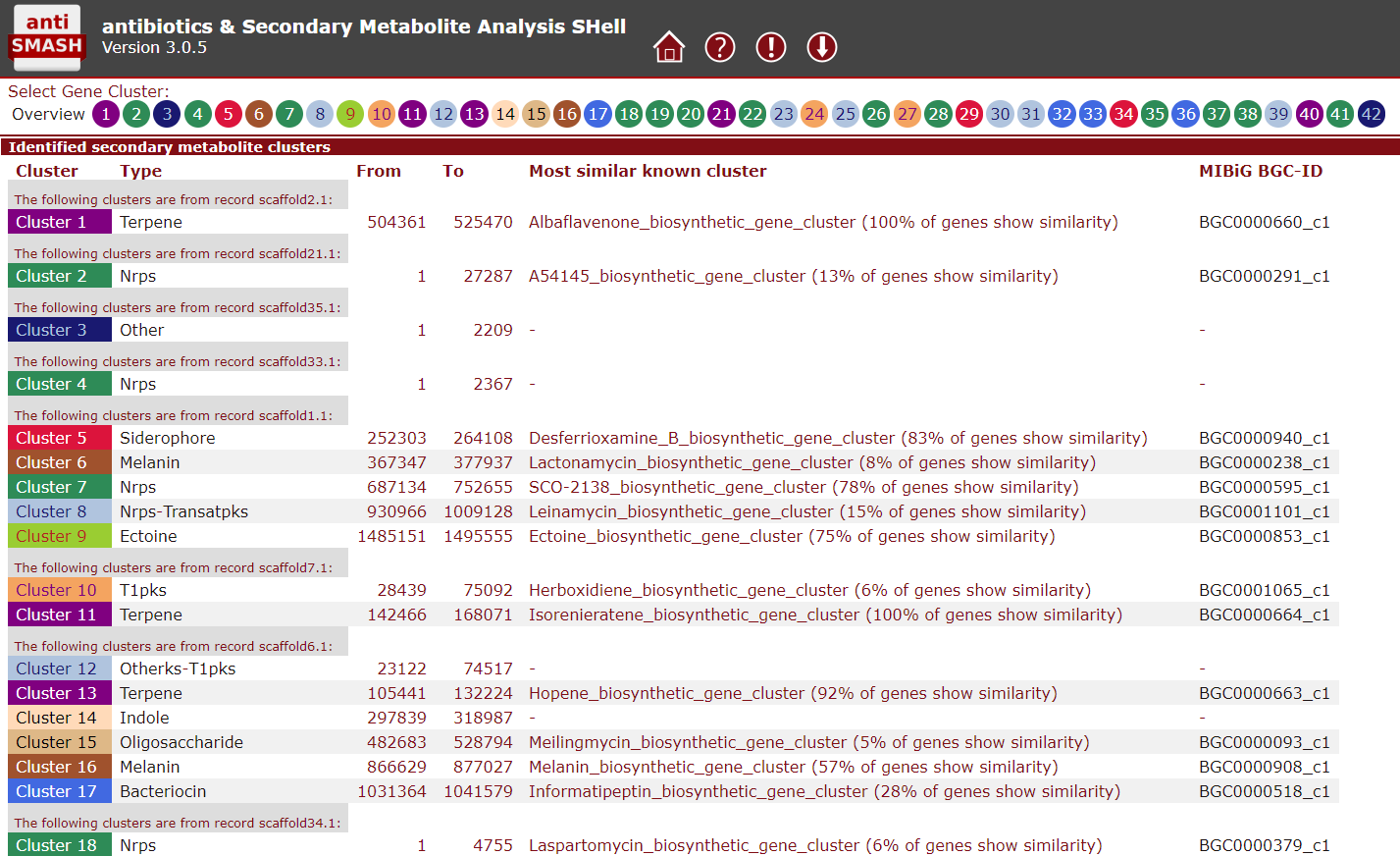
Figure 2. antiSMASH secondary metabolite prediction
antiSMASH is an online tool that can identify putative BGCs based on known core genes. This information is used to identify potentially novel natural products, which can for example be targeted for activation.
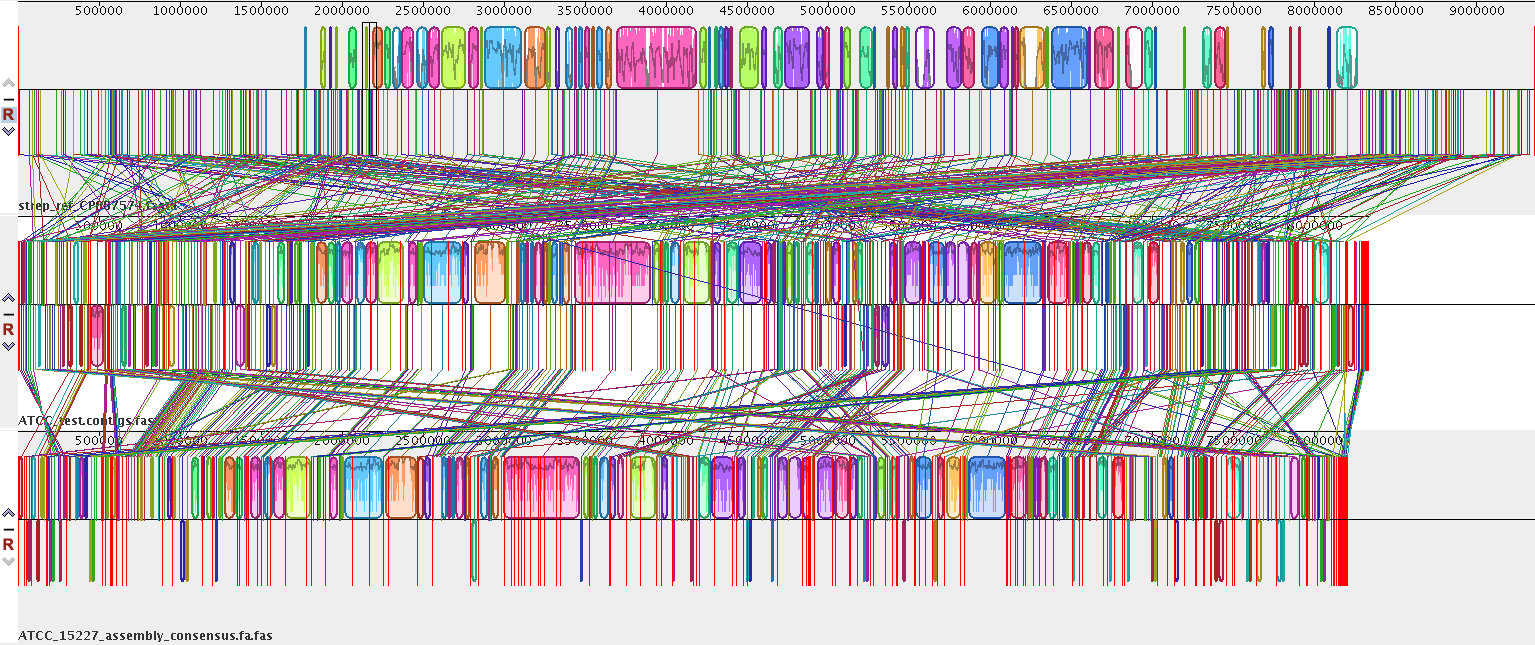
Figure 3. Mauve whole genome comparison
Mauve is a multiple genome aligner that helps us compare whole genomes, which allows us to compare different mutant strains for genetic differences, for example.
Updated: June 2018