Currently, being able to predict the function of an enzyme based solely on its structure is impossible. This is especially true for novel enzymes with unknown function or poor sequence homology to other known sequences. Crystal structures of enzymes can provide valuable insight into the function of an enzyme and elucidate the interactions between enzyme and its substrate, but in order to understand mechanisms of catalysis further experiments are needed. One of the best means to gain insight into structure/function relationships is protein engineering. In cases where the proteins have already been well characterized, protein engineering can be utilized to alter the substrate binding and catalysis to generate novel natural products. Finally, protein engineering is a helpful tool to understand how enzymes evolve to catalyse novel chemistry.
We have traditionally used two strategies for modification of enzymes. The first consists of rational design, whereas a semi-rational approach is utilized in the second strategy. Often both techniques can be combined as they are not mutually exclusive.
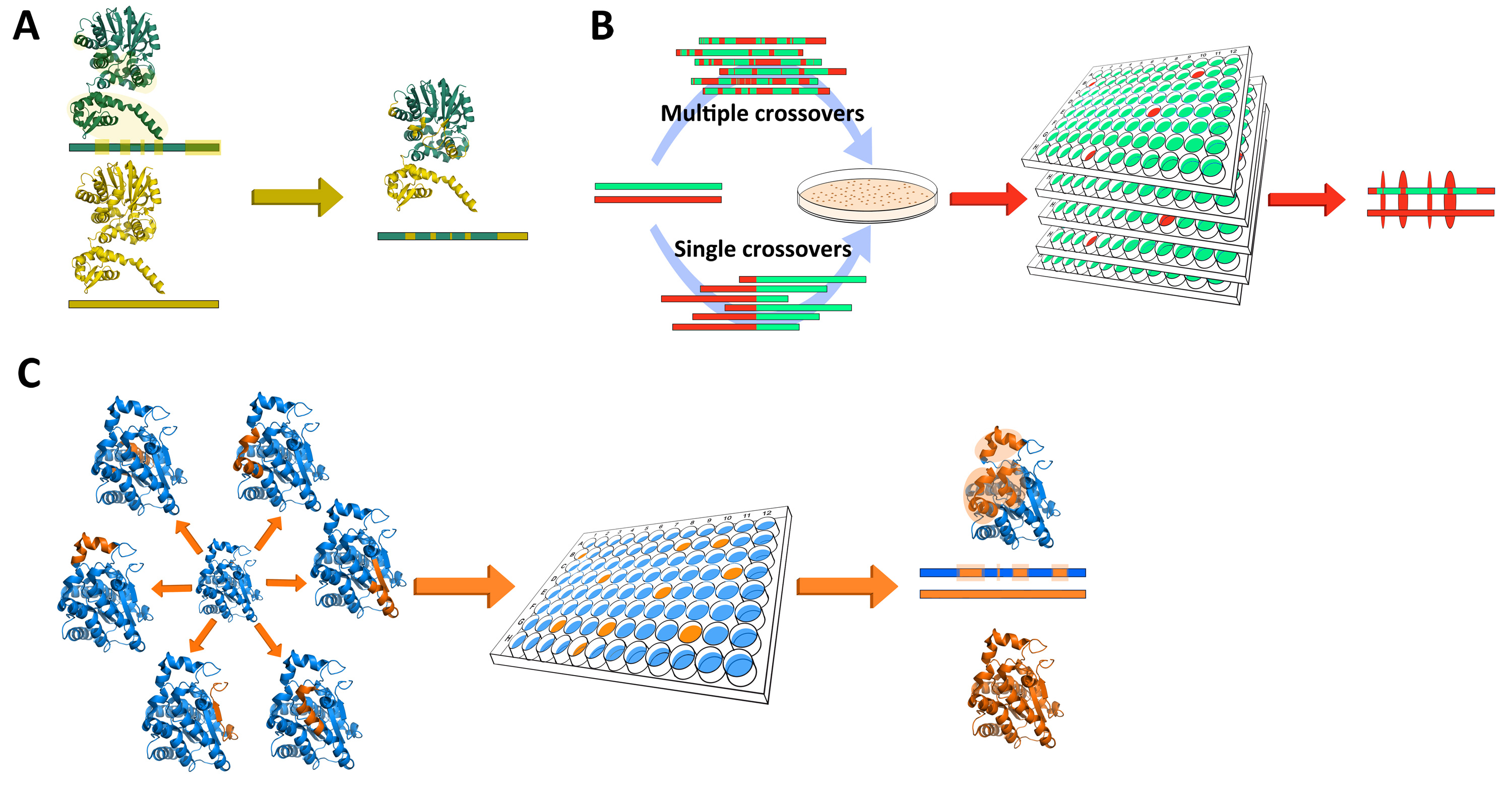
Figure 1. Examples of protein engineering techniques. A: B: In rational design, structural information is used to identify regions of interest that will likely result in the desired outcome. Several distinct regions may be selected and be targeted either individually or in different combinations. The number of key regions identified is usually small. Since the library of chimeras is small, they can usually be tested individually for activity. B: Random chimeragenesis involves generating large sequence libraries through arbitrary sequence combinations. The methodology does not require structural information of the target proteins, but relies heavily on the use of a high-throughput screening system where the enzyme with the desired properties is screened for. The libraries may be constructed through multiple crossovers if the sequence similarity is sufficiently high, but in cases of low sequence similarity single crossovers are still possible. C: Semi-rational design uses structural information to limit the regions of interest as well as the potential sequence space of the library. However, the methodologies still depends on screening methodologies to identify the desired activity.
Rational design:
Often, by tinkering with amino acids in enzymes, valuable information about the amino acids involved in the catalytic mechanism, usually in the active site, can be obtained. By targeting specific amino acids one can gain insight in structural stability, ligand/ cofactor binding, and the catalytic machinery. Using these methods, as well as activity assays and kinetics, we can piece together the mechanisms by which the enzyme can catalyse its reaction.
Random and Semi-rational design:
In some instances, identifying the amino acids involved in catalysis is very challenging, especially when no homologous structures are available or the reaction the enzyme catalyses in unexpected. To solve this issue, a traditional approach involves introducing random mutations throughout the protein sequence (random mutagenesis) to identify crucial amino acids. By creating a large library of mutants it is possible to identify amino acids involved in catalysis by screening the library. However, within the total possible number of sequences possible, only a finite number of sequences lead to stable and catalytically active enzymes. If random sequences are introduced, many of the enzymes generated likely lead to unstable and/or non-functional enzymes.
To better understand and identify specific regions within enzymes that are responsible for catalysis we use as a semi-rational approach named chimeragenesis. By combining stretched of sequence from two or more homologous enzymes, we can identify the areas within a sequence that are vital for the catalytic machinery. Since the amino acids sequences have been preselected by evolution, chimeragenesis typically generates stable and functional enzymes.
Updated: May 2018